No-Code DEL-ML (DNA-Encoded Library Machine Learning) Analysis with Prithvi
Deep Forest Sciences
Rida Irfan, Bharath Ramsundar
05.31.2024
A Brief Overview on DEL Technology
In drug discovery, exploring vast chemical spaces to find potential drug candidates is a major challenge. While traditional techniques like high throughput screening (HTS) have successfully generated numerous leads for development, such methods have major limitations. HTS setups remain costly and are limited to screening about a million compounds.
DNA-encoded libraries (DEL) are a recent development that allow for synthesis and screening of chemical libraries containing billions of small molecule compounds. DELs use water-based combinatorial chemistry and a split-and-pool method to generate diverse compounds. These compounds are screened against target proteins via affinity selection in a single-tube approach, unlike high throughput screening, which requires each compound to be screened individually.
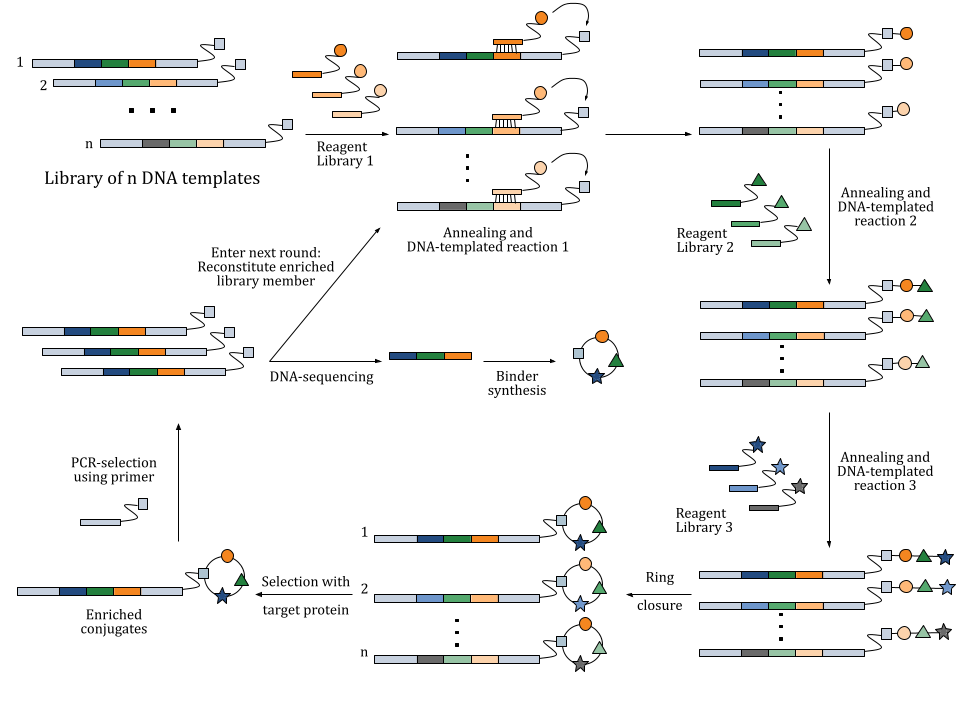
In DELs, oligonucleotides serve as a unique identifier for each compound in the library. These oligonucleotides are short DNA sequences that are chemically linked to small molecule compounds. During the synthesis process, each compound is tagged with a specific oligonucleotide sequence which encodes information about the chemical building blocks from which the compounds were synthesized. When compounds bind to a target protein, the attached oligonucleotides can be rapidly sequenced. This allows researchers to identify successful interactions without the need for separate testing of each compound.
DEL Integration with Machine Learning
DNA encoded libraries have considerable synergy with machine learning methods, since they naturally generate very large scale datasets. A seminal article from Google in 2020 created a lot of interest in the field of DEL-ML by demonstrating how DEL data could be used to train large machine learning models for hit finding. Trained DEL-ML models could be used to rapidly screen large compound libraries to predict activity. Subsequent work has shown that machine learning models like Random Forests (RF) and Graph Convolutional Neural Networks (GCNN) trained on DEL data are effective for predicting the activity of new compounds selected from vendor libraries. The systematic use of DEL-ML holds the potential to significantly reduce the time and cost needed to bring new molecules to market, and the approach has consequently generated significant interest in the industry.
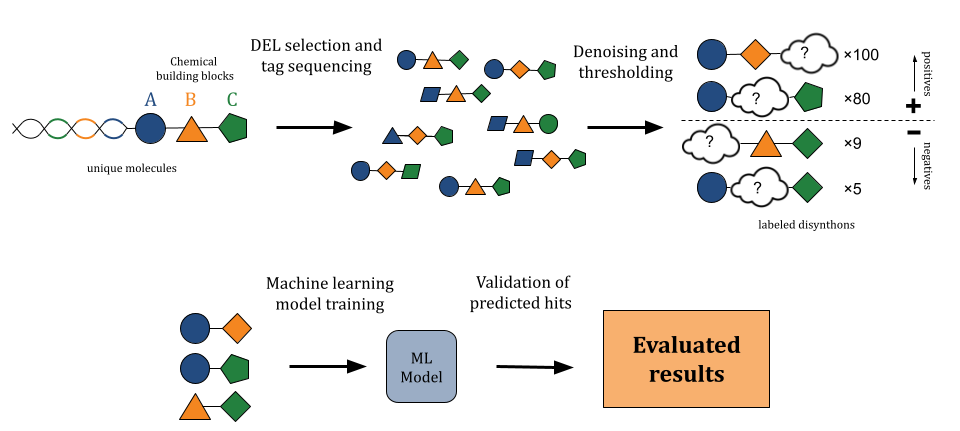
However, computational processing of DEL data remains challenging. There is considerable assay noise in DEL data, which has led to the use of denoising strategies such as “disynthon aggregation,” which looks at subfragments of DEL compounds to lower noise. More recent research has started to explore the use of directly learning on full DEL compounds as well.
No-Code DEL-ML Analysis with Prithvi
Although DEL-ML has found considerable adoption, deploying machine learning solutions for DEL data has remained a difficult technical endeavor, requiring considerable software engineering expertise. To address this need, Deep Forest Sciences has released infrastructure on our Prithvi™ platform to enable biologists and chemists to build no-code DEL-ML models. We anticipate that our technology will enable biologists and chemists to use DEL data systematically in their discovery pipelines.
A Brief Introduction to Prithvi
Prithvi is our AI-powered platform to accelerate small molecule drug discovery with scientific foundation models. Prithvi has the ability to
- Perform structural analysis of targets and identify potential binding sites for a more targeted design process.
- Build models based on early assay or patent data for use in large-scale virtual screens.
- Construct active learning pipelines to identify and confirm high-quality hits.
- Guide retrosynthetic analysis.
- Suggest modifications to increase the potency, selectivity, and safety of hits.
To learn more about Prithvi, check out our earlier blog post. In this blog post, we will discuss the DEL-ML capabilities of Prithvi.
No-Code DEL-ML
Prithvi’s DEL pipeline offers no-code machine learning tools designed to model, analyze, and virtually screen small molecule compounds. Prithvi streamlines the entire DEL-ML workflow, from visualizing and denoising DEL data to training models and identifying hits. Prithvi’s no-code infrastructure allows scientists to focus more on discovering new hits and less on the technical complexities of machine learning pipelines.
To perform Prithvi DEL-ML analysis, users can run the following no-code workflow:
- Visualize DEL data.
- Denoise DEL data.
- Featurize denoised data.
- Split featurized data.
- Optimize hyperparameters.
- Train models to predict hits.
- Evaluate trained models.
- Run inference on vendor catalogs.
- Threshold hits and cluster for chemical diversity.
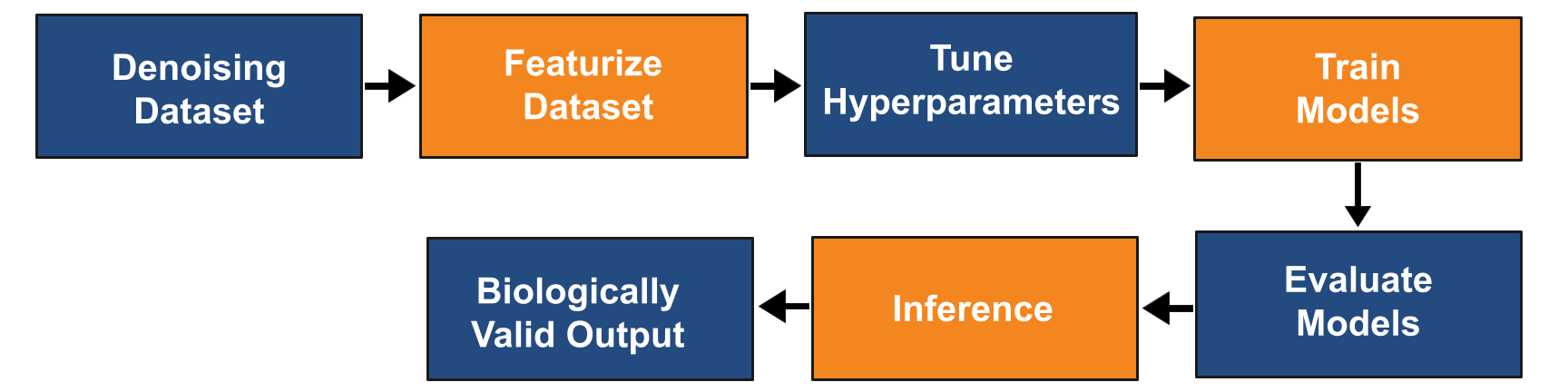
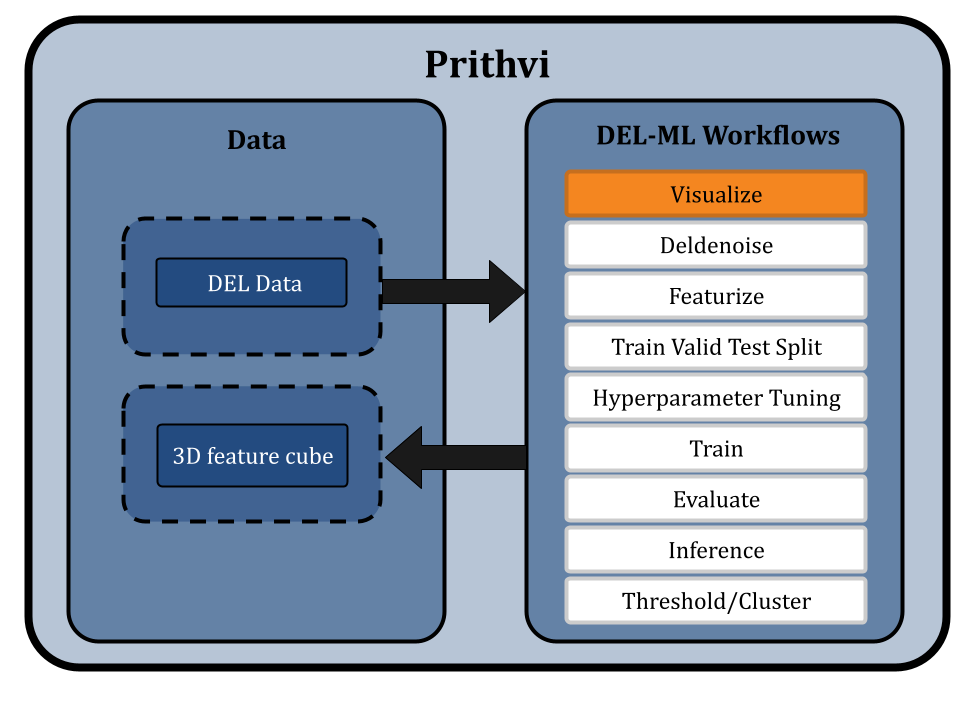
This workflow consists of a series of no-code primitives which can be invoked by users directly from Prithvi’s web user interface (UI). Advanced users can save workflows as reproducible scripts which can be invoked from the UI or alternatively from a Python API. The workflows have been tested on live DEL projects at large scale with existing partners.
Let’s discuss each workflow in the Prithvi DEL-ML pipeline.
Visualize DEL Data
The Visualize primitive in the Prithvi DEL pipeline renders DEL data in a 3D “feature cube.”. This tool provides users a comprehensive view of DEL data, and helps identify “line” and “plane” features (one or two conjoined DEL building block fragments which are commonly represented in hits). Each axis represents a building block and color-coded counts help identify hits.
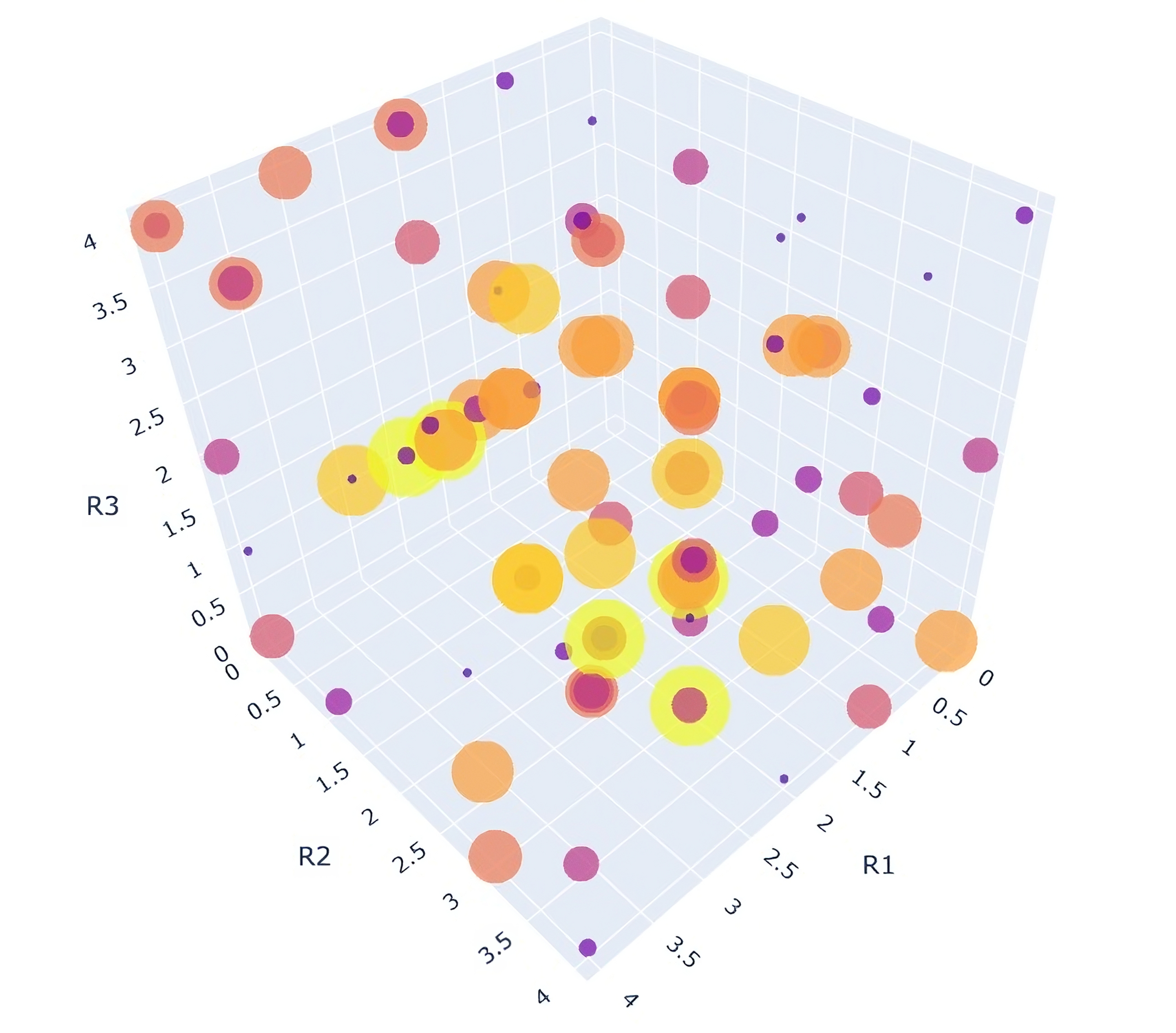
Denoising DEL Data
Input DEL data is noisy due to noise in sequencing. A common denoising strategy is to use aggregated sequencing counts based on shared “disynthon” representations. For instance, in a three-cycle library of the form A−B−C, sums aggregating counts for the A−B, A−C, and B−C disynthons can be generated to lower noise.
Suitable cutoffs can be set for aggregated reads (sequence counts) to ensure that small sequence counts don't influence the balance of the training data. The following formula has been proposed in the literature as a strategy for converting sequence counts into enrichment scores,
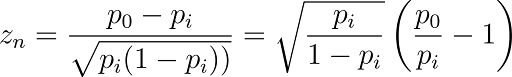
This normalized z-score calculation approximates the DNA sequencing process as random sampling with replacement using a binomial distribution. It quantifies the enrichment level of an observed count compared to the expected count, factoring in the sequencing read depth and library sizes. The average of these scores sets a threshold that categorizes each disynthon pair as a "hit" or "not hit."
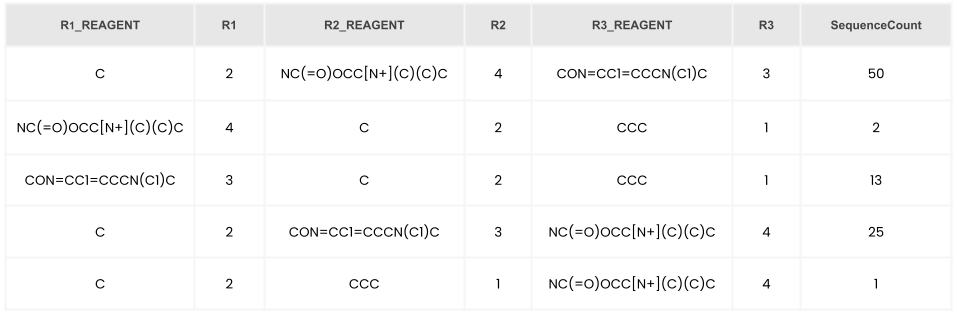
Prithvi’s Deldenoise primitive implements denoising strategies following these and other best practices from the literature to enable users to easily convert raw assay data into a format suitable for machine learning.

Featurizing Denoised Data
The Featurize primitive in Prithvi is used to featurize the denoised DEL data. (Here “featurization” denotes the process of converting inputs into a form suitable for machine learning.) This step involves transforming denoised outputs, such as disynthons, which can be represented as pairs of SMILES, into a disjoint union of graph structures (for GCNNs) or fingerprint vectors (for RFs).
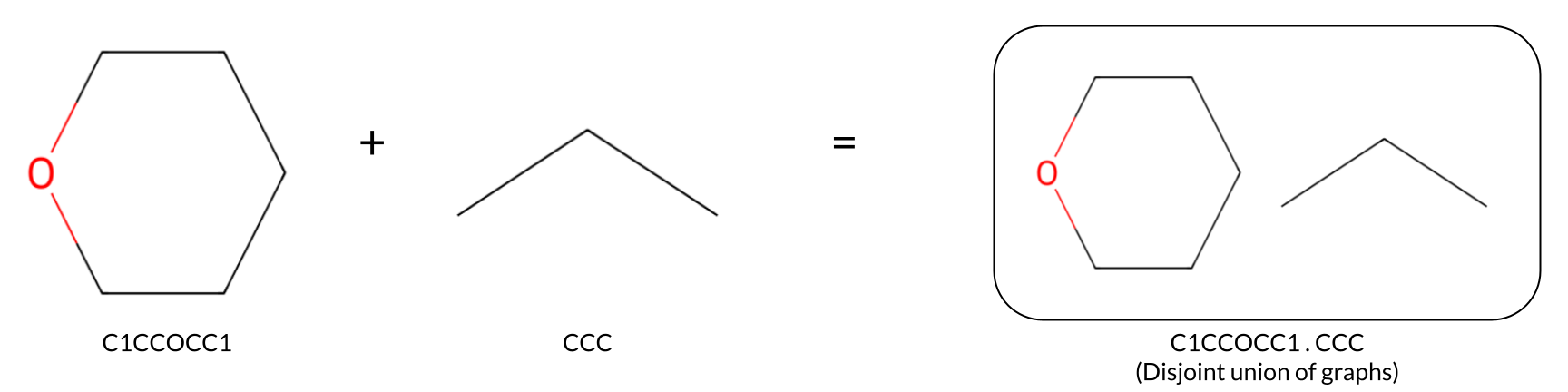

Split Featurized Data
The featurized data is split into training, validation, and test splits using the Split primitive. In scaffold splitting, the data is split into training and test sets such that the molecular scaffolds in each group are as distant from the other as possible. The result is that every single molecule in the test set has a scaffold unique from the training set, and is structurally dissimilar from the training set. Ensuring the test set is maximally distant from the training set helps test models for generalizability and provides a better measure of their performance on new molecules.
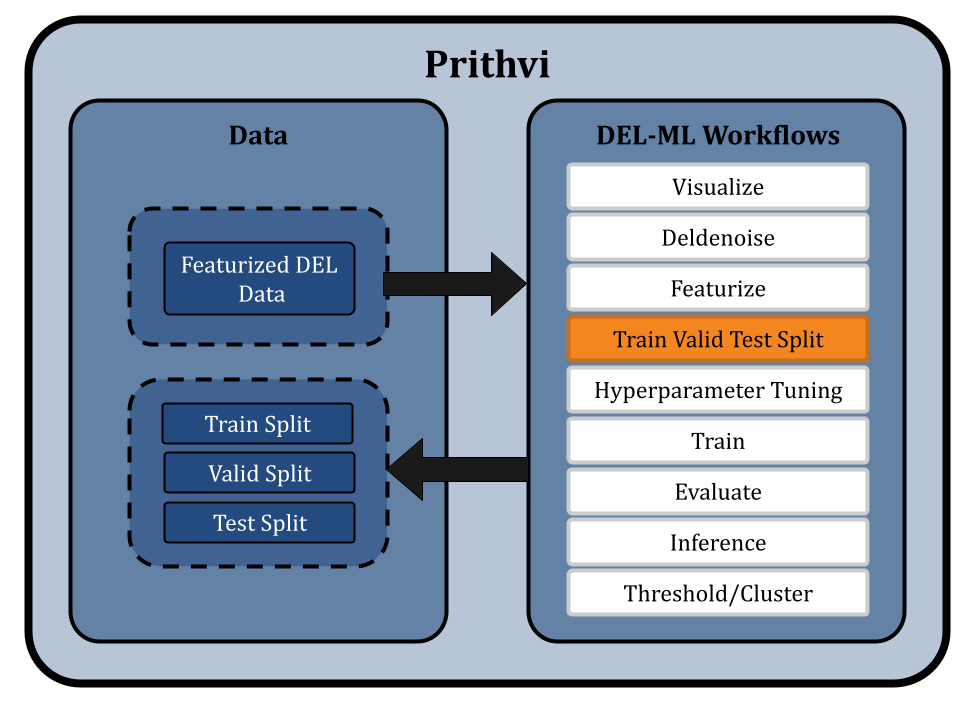
Optimizing Hyperparameters
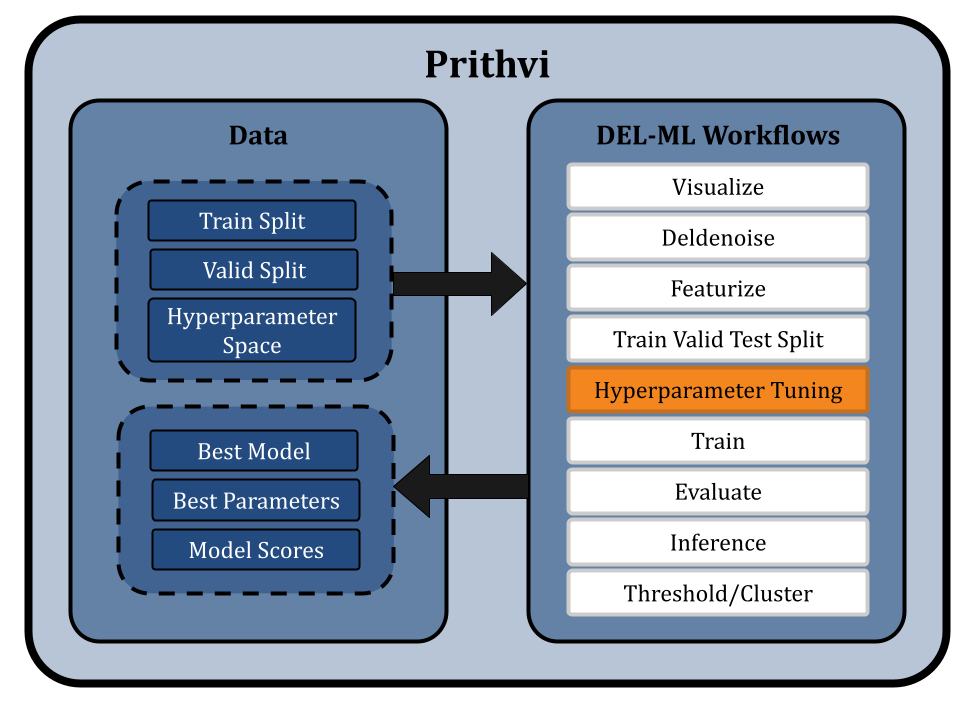
The Hyperparameter Tuning primitive in Prithvi uses the training and validation splits of the featurized DEL data to train several representative machine learning models, like GCNs and RFs, using different combinations of hyperparameters from the provided hyperparameter space. Prithvi employs a hyperparameter search technique, which systematically searches through a specified hyperparameter space and evaluates the performance of the model for each combination of hyperparameters.
Hyperparameter tuning is crucial in the context of DEL data due to the large chemical diversity and inherent noise in the library. The output of this workflow provides the best model, the best hyperparameters, and a collection of scores for all evaluated models for hit predictions.
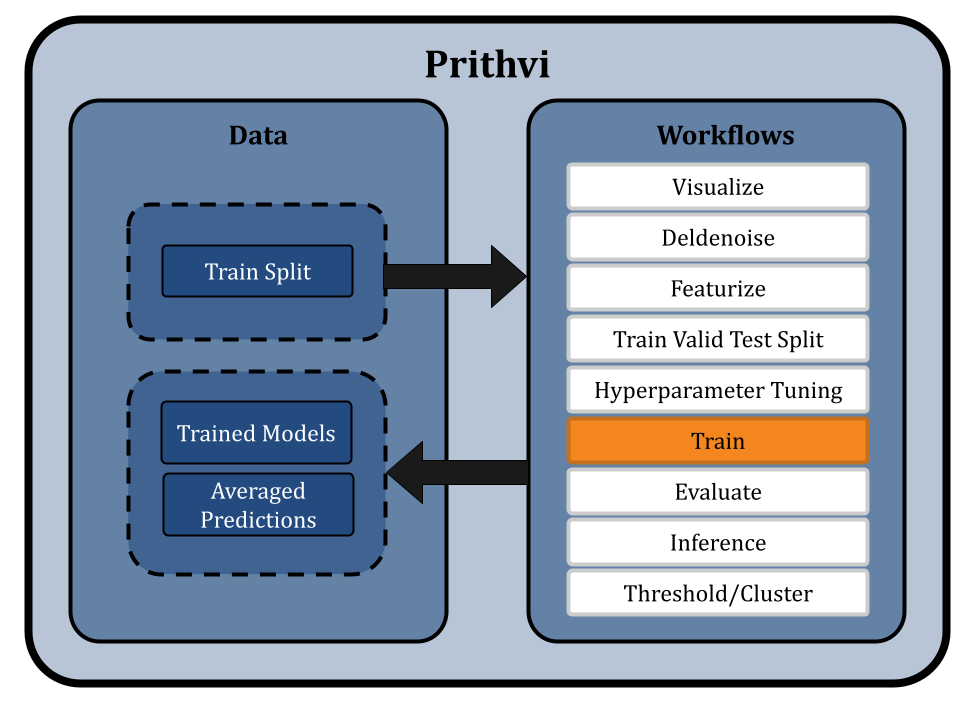
Training Models
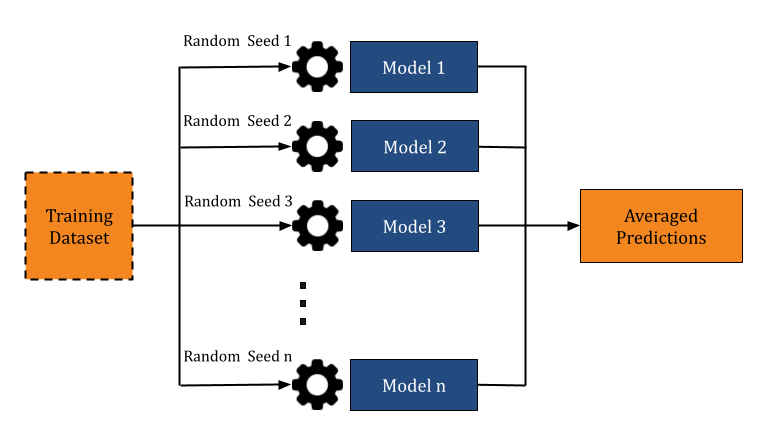
We use the Train primitive to train an ensemble of GCN and RF models to enhance the accuracy of hit predictions from DEL data. Prithvi supports no-code construction of ensembles, which are collections of multiple copies of a base model each trained on the same training data but with different random seeds. The predictions from these individual models are then averaged to generate a final robust result.
Because ensemble models combine predictions from multiple models, they improve accuracy by reducing the impact of individual model biases. This approach leads to better hit identification by minimizing false positives and offering consistent predictions across diverse DEL datasets.
Evaluating Trained Models
Prithvi’s Evaluate primitive is used to evaluate the performance of trained models using metrics such as ROC-AUC, which measures the area under the Receiver Operating Characteristic curve. This evaluation can be run on the train, valid, and test splits as desired. Prithvi computes these statistics for each model and saves the results in a CSV file.
Run Inference on Vendor Catalogs
The Infer primitive is used to generate hit predictions for SMILES on specified vendor catalogs using trained ensembles of models.
Threshold Hits and Cluster for Chemical Diversity
Users can use the Threshold primitives to select the top 1% (or any other desired percentage) of predictions as hits. Once thresholds are identified from the inference results, users can run the Cluster primitive to cluster these hits and select representative top hits from cluster centers.
Conclusion and Takeaways
DNA-encoded library (DEL) technology can significantly accelerate the discovery process by enabling systematic hit discovery. Prithvi™ provides a powerful no-code tool for DEL-ML analysis, enabling biologists and chemists to process DEL data without needing sophisticated Python pipelines. Prithvi streamlines the entire process of modeling, analysis, and screening of small molecule compounds into a series of no-code primitives, which can be modularly composed and modified as users require. Each stage of Prithvi’s DEL pipeline, from data visualization to advanced hit prediction, is designed to optimize the discovery and evaluation of potential drug candidates, allowing scientists to focus more on discovery and less on technical complexities.
About Deep Forest Sciences
Deep Forest Sciences’ no-code AI Prithvi™ toolchain accelerates small-molecule drug discovery efforts. Deep Forest Sciences also leads the development of open-source DeepChem framework, and emphasizes supporting open source and open science as fundamental parts of our mission and values. Partner with us to apply our foundational no-code AI technology to hard real-world problems in small molecule drug discovery.
Email us at partnerships@deepforestsci.com to learn more!